In our daily work as data scientists, we deal with a lot with tabular data, also called DataFrames. At Blue Yonder, we use Pandas DataFrames to analyze and transform the data we need. One of our main challenges when we integrate new systems into our software landscape is how we can pass data between it and Pandas. When data is persisted in a file-based storage, we utilize the Apache Parquet format.
Choosing a file format
For database integration, we use turbodbc, which we developed and posted to GitHub, as a high-performance access layer. However, the current file formats supported by Pandas are either very efficient or very portable; Never both. To keep our data scientists productive with the tools they choose, we need a file format that excels at both.
The most portable format for DataFrames is CSV. Due to its simplicity, every tool has some way to load CSV files. However CSV stores data row-wise as text without a schema. While this makes parsing CPU intensive, it also means that every parser may interpret the data differently. Although there were several attempts to annotate CSV files with basic schemas, these approaches never gained wide support. On the other hand, through HDF5 and msgpack, Pandas has two very fast data formats with a schema for columns. There is wider support for HDF5 in other numerical tools, but msgpack files generated by Pandas cannot be read by any other tool and some cannot even be read between different Pandas versions. Looking at the zoo of data analytics tools housed in the Apache Foundation, it’s hard to find support for one of these file formats in this ecosystem.
Tools with an Apache prefix and their related ecosystem always list Apache Parquet as one of their preferred formats. While some tools have custom file formats, Parquet is universally supported and is often a requirement for effective use of their tool. If you look at Apache Spark’s tutorial for the DataFrame API, they start with reading basic JSON or txt but switch to Parquet as the default format for their DataFrame storage as it is the most efficient. In this respect, Pandas has long been an outlier as it had not offered support for operating with files in the Parquet format. Nowadays, reading or writing Parquet files in Pandas is possible through the PyArrow library.
Apache Parquet is a columnar format with support for nested data (a superset of DataFrames). It is based on the record shredding and assembly algorithm described in the Dremel paper. For each column, very efficient encoding and compression schemes are applied. In its core, it was designed to be a portable between the various data processing frameworks in the Hadoop ecosystem. Now its reach is much further. There are also several hints and structures integrated into the format so that queries on Parquet files can be executed efficiently and large parts of the query conditions can be pushed into the I/O layer. This means that only the necessary chunks of the data is read and materialized from disk.
Reading Apache Parquet files with Pandas
The code below shows that operating with files in the Parquet format is like any other file format in Pandas. Support is provided through the pyarrow package, which can be installed via conda or pip.
import pyarrow.parquet as pq
df = pq.read_table('<filename>').to_pandas()
# Only read a subset of the columns
df = pq.read_table('<filename>', columns=['A', 'B']).to_pandas()
The Parquet implementation itself is purely in C++ and has no knowledge of Python or Pandas. It provides its output as an Arrow table and the pyarrow library then handles the conversion from Arrow to Pandas through the to_pandas() call. Although this may sound like a significant overhead, Wes McKinney has run benchmarks showing that this conversion is really fast. In a future Pandas version 2.0, this overhead will be completely gone.
Reading in subsets of columns is a typical data science task. Due to its columnar nature, Parquet allows for efficient reading of data into memory by providing the columns argument. In this case, the metadata of the Parquet files is first parsed and then the positions of the requested columns in the file are retrieved. Finally, only the section from the file containing these columns are loaded, reducing the amount of data that needs to be loaded from disk, passed through memory and CPU caches. This also saves CPU time as there is no need to deserialize the data for the unwanted columns.
Writing Apache Parquet files with Pandas
import pyarrow as pa
import pyarrow.parquet as pq
table = pa.Table.from_pandas(data_frame, timestamps_to_ms=True)
pq.write_table(table, '<filename>')
Writing a Pandas DataFrame into a Parquet file is equally simple, though one caveat to mind is the parameter timestamps_to_ms=True: This tells the PyArrow library to convert all timestamps from nanosecond precision to millisecond precision as Pandas only supports nanoseconds timestamps and deprecates the (kind of special) nanosecond precision timestamp in Parquet.
In contrast to the read path, the Pandas to Arrow conversion is faster as it allows us to generate most columns in the Arrow table without copying from the columns in the Pandas DataFrame.
Benchmarks
Filesystem storage formats can excel in different categories. For a file format and its Pandas implementation, there are two properties that are of relevance to us: read performance and storage size. These two also directly translate into operational costs (“How much do we have to pay for storage each month?”, “How long do our Data Scientists idle while waiting for the data to load?”). To illustrate how Apache Parquet performs, we have used the New York Taxi & Limousine Commission Trip Data dataset. This DataFrame consists mostly of integer and floating point columns, a single boolean and two date columns.
Reference Data
As the basis for the benchmarks, we use the time range from January 2015 to June 2016 to analyze the compression ratio each format achieves in comparison to the original baseline data of 31.8 GB. For measuring the read performance, only the first file for January 2015 was used. The following code was used to convert the initial CSV files to the respective file formats.
df = pd.read_csv(key, parse_dates=['tpep_pickup_datetime', 'tpep_dropoff_datetime'])
df['store_and_fwd_flag'] = df['store_and_fwd_flag'] == 'Y'
# CSV with GZIP compression
df.to_csv(filename, index=False, compression='gzip')
# msgpack uncompressed
df.to_msgpack(filename)
# msgpack with zlib compression
df.to_msgpack(filename, compress='zlib')
# HDF5 uncompressed
df.to_hdf(filename, 'data', mode='w')
# HDF5 with zlib compression
df.to_hdf(filename, 'data', mode='w', complib='zlib')
# HDF5 with blosc compression
df.to_hdf(filename, 'data', mode='w', complib='blosc')
# Convert DataFrame to Apache Arrow Table
table = pa.Table.from_pandas(df, timestamps_to_ms=True)
# Parquet uncompressed
pq.write_table(table, pq_name, compression='NONE')
# Parquet with Snappy compression
pq.write_table(table, pq_name)
# Parquet with GZIP compression
pq.write_table(table, pq_name, compression='GZIP')
# Parquet with Brotli compression
pq.write_table(table, pq_name, compression='BROTLI')
Storage Size
Reducing the file size has a significant impact on your storage bill. It also translates into large time savings when your data is transferred over network. While files on an SSD have transfer rates that are in the range of gigabytes per second, network bandwidth is more in the region of 20 to 60 megabytes per second . Thus, the time data needs to reach the worker node might greatly outweigh the time it takes to decode the resulting DataFrame from the file format.
One of the main advantages of Parquet is its storage efficiency. In comparison to other formats, universal compression algorithms (e.g. gzip) are applied on the data and type-specific encoding techniques are used on each column to minimize the storage size. The most common encodings are dictionary encoding and run-length encoding, which excel on repetitive data. In contrast to universal compression, these techniques are limited to data of the same type but are lightweight and do not need much CPU processing power. Simply by using the encodings on the data, Parquet files only have a fifth of the size of the original (UTF-8 encoded) CSVs.
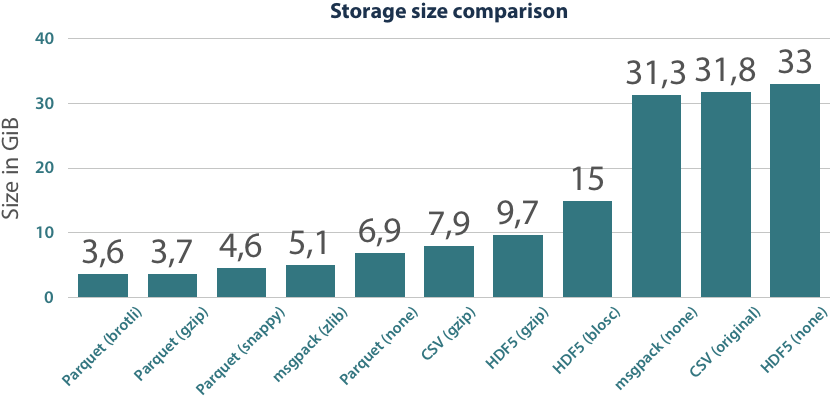
Using universal compression codecs, we can save another factor of two in the size of Parquet files. A speciality of the Parquet format is that the compression is applied to individual segments of a file, not globally. While this has a small impact on the achieved compression ratios, it is very beneficial in the read and query capabilities of this format. The only file format that can reach similar compression ratios as Parquet does is msgpack. While it has the same binary data and compression codec as HDF5+gzip, its internal layout can be more beneficial to the compression algorithm and so it achieves this ratio.
Read Performance
Another aspect of a file format is how quickly you can deserialize it into a DataFrame. The your I/O subsystem (e.g. hard drive, network storage, etc.) is, the more relevant this serialization time becomes. For slow I/O deserialization, the execution mostly waits for the data to be loaded from disk, while the deserialization itself can be the limiting factor in faster subsystems . To remove the impact of the I/O subsystem, we ran the benchmarks on files that resided in memory.
We have always measured the time it takes to get from the serialized bytes into a Pandas DataFrame. This also included the timings for to_pandas() in the measurements that we did in the Parquet conversion. The benchmarks were run on an Intel(R) Xeon(R) CPU E5-2680 v2 @ 2.80GHz.
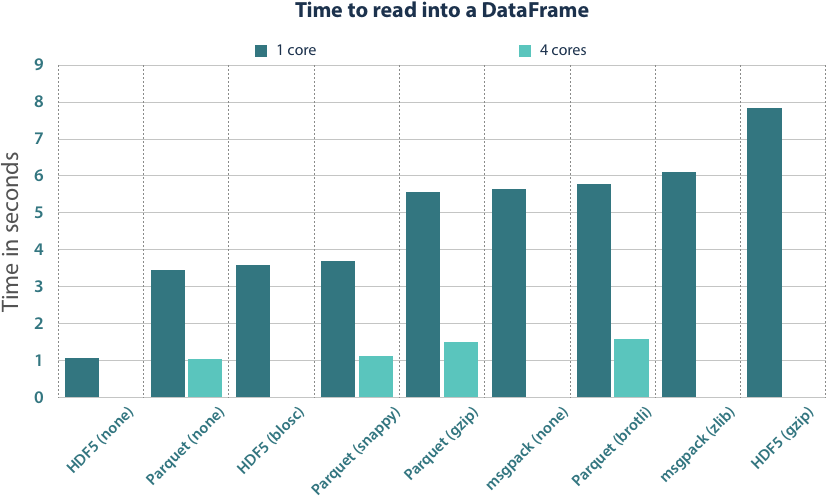
Looking at the results, HDF5 is the clear winner in single thread performance. The data here is laid out just like it should reside in memory, so the file loading is merely allocating the memory regions and copying the data into it. In Parquet files, columns are often encoded so that they cannot be directly copied, but need to be decoded and uncompressed. Thus, it is unrealistic to achieve the same single-thread performance.
Still, Apache Parquet can achieve the same read performance level as HDF5. The format is structured in such a fashion that you can independently read columns and even chunks of them in parallel. We added the time measurements for 4 cores, which are realistic for workstation and server machines. With parallelization, we can spread the CPU workload better across the individual cores, but it also provides a better utilization of the memory bus that cannot be fully exploited by a single core in current machines. With 4 cores, we are then able to come in the same timing region as the ultra-fast plain HDF5 files.
Another aspect of a file format is its write performance, but it is less important than the read performance, especially if you read your file several times.
Read and Storage performance combined
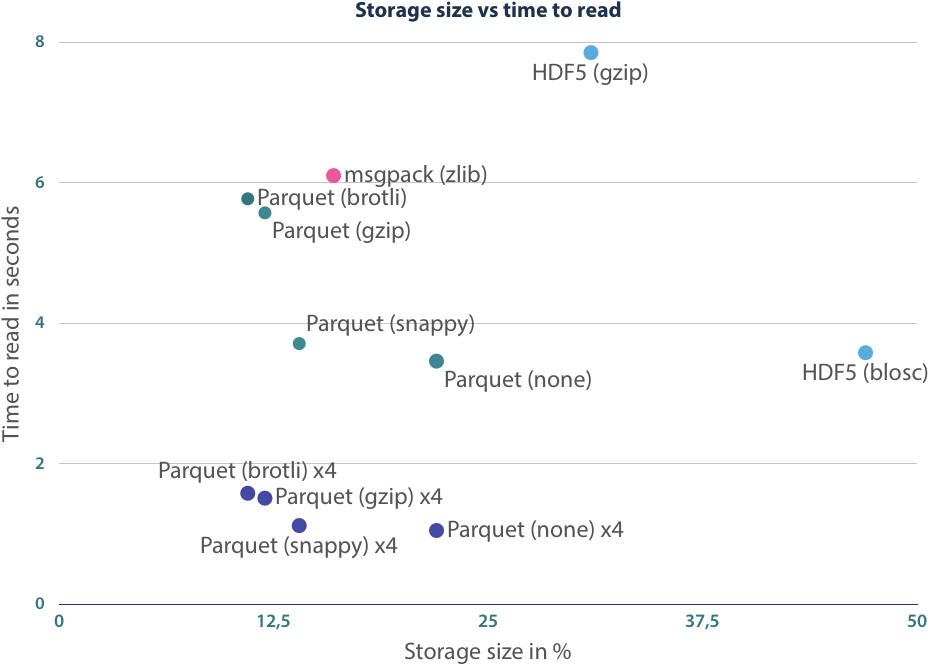
With the benchmarks above, we have shown that we can get decent performance using Apache Parquet files, but the user must decide which configuration is the best. The trade-off here fully depends on the available I/O bandwidth. If a high throughput is provided, choose either no compression or the very fast snappy option. For the low throughput scenario, Brotli compression is your best bet.
Get in action
With official Python/Pandas support for Apache Parquet you can boost your data science experience with a simple pip install . Check out the documentation, report issues or dive into the Apache Arrow / Apache Parquet C++ code and contribute any missing features or bugfixes.