Run-length Encoding for Pandas
Pandas is a very powerful tool for data science. It allows data processing and inspection in a convenient manner. The DataFrames in use often contain flat data produced from normalized data sources. Imagine the following (simulated) weather dataset:
| date | month | year | city | country | avg temp | rain | mood |
|---|---|---|---|---|---|---|---|
| 2000-01-01 | 1 | 2000 | Berlin | DE | 12.4 | False | ok |
| 2000-01-02 | 1 | 2000 | Berlin | DE | 4.0 | False | ok |
| 2000-01-03 | 1 | 2000 | Berlin | DE | 7.2 | False | great |
| 2000-01-04 | 1 | 2000 | Berlin | DE | 8.4 | False | ok |
| 2000-01-05 | 1 | 2000 | Berlin | DE | 6.4 | False | ok |
| 2000-01-06 | 1 | 2000 | Berlin | DE | 4.4 | False | ok |
| … | … | … | … | … | … | … | … |
| 2005-12-31 | 12 | 2005 | Berlin | DE | -3.1 | False | ok |
| 2000-01-01 | 1 | 2000 | Hamburg | DE | 9.1 | False | ok |
| 2000-01-02 | 1 | 2000 | Hamburg | DE | 8.2 | False | ok |
| … | … | … | … | … | … | … | … |
| 2005-12-31 | 12 | 2005 | Hamburg | DE | -2.1 | False | ok |
Especially attribute columns like month, year, city, and country contain loads of duplicated data, but are
important to analyze and plot the data as well as as an input for machine learning models. For example, looking at the
city column, we can see that the same value "Berlin" stored consecutively multiple times before the entries for
"Hamburg" occur. Instead of storing each entry, it would for sure be smarter to just store a city together with a
number of times it is repeated, e.g. “Berlin - 1000 times, Hamburg - 500 times, …”. This is what
Run-length encoding does. It can compress that data without
precision-loss while at the same time it can also be faster to process then the uncompressed data. In our benchmark we
see a decrease in memory consumption by over 200x while being over 50 times faster.
rle-array, which we now open source, implements this technique for the
well-known Pandas library by using the new but powerful
ExtensionArray. The usage
is quite simple and the integration in pandas works well:
# get a DataFrame like the example above
df = ...
# make rle-array available in pandas by importing the package
import rle_array
# convert to run-length encoded
df = df.astype({
"city": "RLEDtype[object]",
"country": "RLEDtype[object]",
"month": "RLEDtype[int8]",
"mood": "RLEDtype[object]",
"rain": "RLEDtype[bool]",
"year": "RLEDtype[int16]",
})
# work with the DataFrame as you would normally do
df["happy"] = (
(df["month"] > 4)
& (df["month"] < 8)
& (df["city"] == "Hamburg")
)
print(df["happy"].sum())
Make sure your DataFrame is sorted, otherwise rle-array will not work efficiently.
Encoding
Run-length encoding is a simple yet powerful technique. Instead of storing array data element-wise, it first identifies so called “runs” — consecutive elements of the array where the same value is stored. For each run, it then just keeps its value and length:

Pandas requires us to be able to do quick random access, e.g. for sorting and group-by operations. Instead of the actual run-lengths we store the end positions of each run (this is the cumulative sum of the lengths):

This way, we can use binary search to implement random access.
Performance and Memory Usage
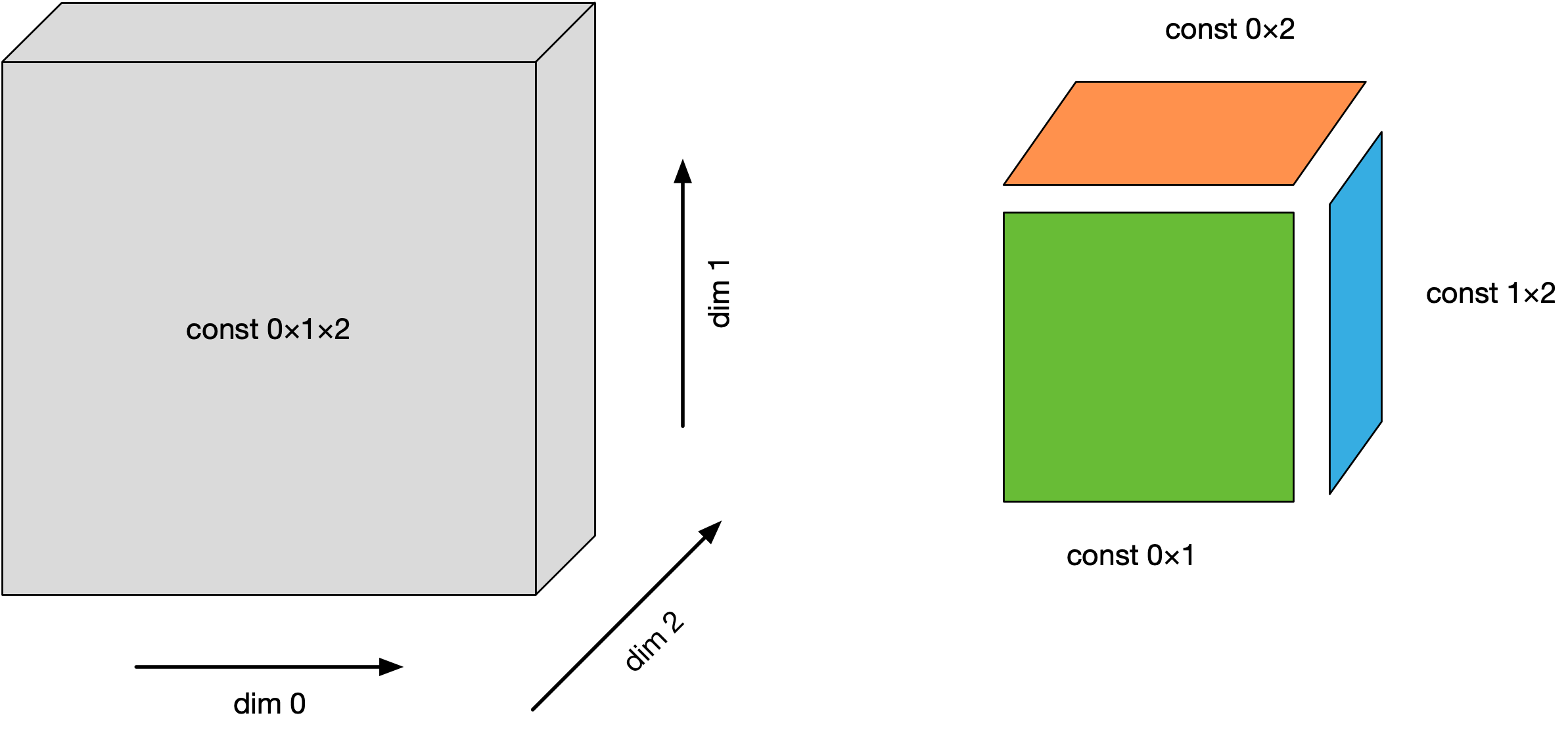
To illustrate the performance of rle-array, we simulate data in form of a 3-dimensional cube with increasing edge length. Every cell of the cube makes up a DataFrame row, similar to what you would expect by pulling data from a normalized database. The DataFrame looks similar to this (edge length is 2):
const_0_1 |
const_0_1_2 |
const_0_2 |
const_1_2 |
dim_0 |
dim_1 |
dim_2 |
|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 4 | 2 | 0 | 1 | 0 | 0 |
| 1 | 2 | 0 | 2 | 0 | 1 | 0 |
| 3 | 6 | 2 | 2 | 1 | 1 | 0 |
| 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| 2 | 5 | 3 | 1 | 1 | 0 | 1 |
| 1 | 3 | 1 | 3 | 0 | 1 | 1 |
| 3 | 7 | 3 | 3 | 1 | 1 | 1 |
This is very similar to the introduction example which has the 2 dimensions “location” and “time”. The columns are created as followed:
dim_{0,1,2}: This symbolizes the dimensions of the cube. Think of attributes like “location” and “time”. There arelengthunique values.const_X_Y: This represents values that are a crossproduct of the mentioned 2 dimensions. Think of attributes like “weather” (which is a crossproduct of “location” and “time”). There arelength^2unique values.const_0_1_2: These are the inner cells of the cube. There arelength^3unique values, as many as there are rows in the DataFrame.
The whole setup can also be visualized:
You can generate the same data using
rle_array.testing.generate_test_dataframe.
With growing edge size of the cube, the number of rows grows in a polynomial fashion:
In the following measurements, the const_1_2 is used. Assuming that the column is stored using int64, the
compression over the baseline grows linearly with the edge size of the cube:
Apart from a massive reduction in memory usage, many tasks can also be processed faster. For this benchmark, we choose the following 3:
- add:
df["const_1_2"] + df["const_1_2"] - compare:
df["const_1_2"] == df["const_1_2"] - groupby:
df.groupby("dim_1")["const_1_2"].sum() - sum:
df["const_1_2"].sum()
For very small cube sizes, rle-array does not provide any performance benefits, but quickly (at a edge length of 100) it
starts to outperform the uncompressed baseline, providing over 50 times speedup. This is because many operations in
rle-array are implemented using Numba. However, for operations like group-by the current
Pandas 0.25.x has some performance issues (for the upcoming Pandas 1.0, it looks like we are at least on par with the
baseline):
Summarizing this we can say, the performance depends on the length of the runs — longer runs give better compression and performance; and that rle-array works great for simple operations but has issues with group-by operations like agg and transform.
Related Techniques
Run-length encoding is not the only way to compress data in a sensible way and sometimes other techniques may be better suited for the concrete use case. Here is a short illustration about a selection of techniques and readers are encouraged to try these and others.
Dictionary Encoding
Dictionary encoding replaces the actual payload data with a mapping. The trick is that mapped values can often be more memory-efficient, especially when the original data is very long (e.g. for strings) and are repeated multiple times:

This is what Pandas Categoricals implement. For data-at-rest, this is implemented by Apache Parquet as well.
Note that this is often combined with other techniques and then can also be efficient for data like floating points, since in many cases like accounting the actual number of unique values is small.
Data Types
Pandas lets you not only choose the semantics of a data type (e.g. integer, floats, booleans), but in some cases also the size of it:
| Semantics | Types |
|---|---|
| Signed Integer | int8, int16, int32, int64 |
| Unsigned Integer | uint8, uint16, uint32, uint64 |
| Floats | float16, float32, float64, float128 |
This distinction between semantics and data size is also made by the Kartothek type system.
Here is how this looks like in memory (for big endian machines):
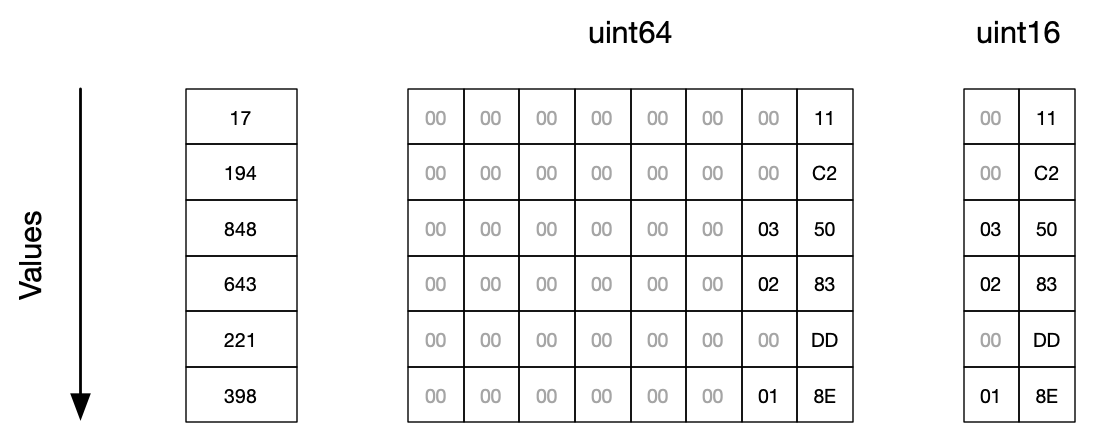
In this example, we can easily use 16 bits per element instead of 64, resulting in a 75% memory reduction.
While using the right data type can lead to significant memory reductions and speedups (float16 and float128 being a
noticeable exceptions due to the lacking hardware support on most CPUs), it also has
some unexpected side effects.
Bit-packing
Bit-packing is similar to Data Types, but allows to create types with non-standard width:
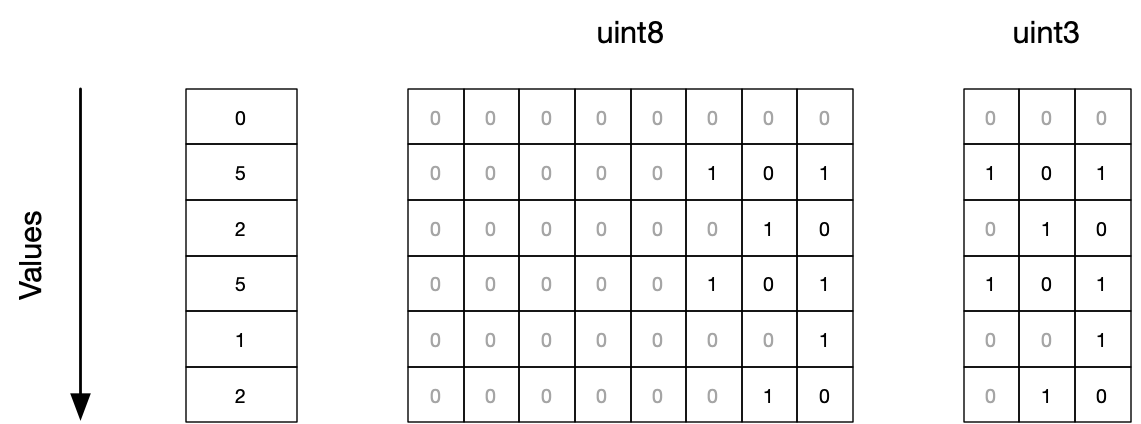
The advantage is that you can save even more memory, but it comes with heavy performance penalties, since CPUs cannot read unaligned data that efficiently. In some cases however, it can be even faster due to the saved memory bandwidth, especially for booleans that normally occupy 8 bits per element.
Bit-packing is — in combination with run-length-encoding — even supported by Apache Parquet.
Sparse Data
Often we find columns in our DataFrames where information only occurs for a very small amount of rows. In that case, it is often more efficient to explicitly store and look-up these few cases — e.g. by using a HashTable — than using a simple array:

This is what Pandas SparseArray implements. Note
that the default value does not need to be 0, but can be an arbitrary element. One downside of sparse arrays is that
you can only have a single default value.
Outlook
For many operations (like
fillna),
we are currently using the default provided by pandas. These implementations however
decompress the run-length encoded data and perform the operation mostly on an NumPy array. While
this is functionally correct, we would like to stay in the compressed format and implement our own specialized version
of it. This will be done step-by-step in future releases.
Since rle-array is (to my knowledge) apart from SparseArray the first compressed ExtensionArray in the pandas
ecosystem, we will likely face some performance bugs and integration issues. We will collaborate with the pandas
community and hope to find a solution. To improve this, we will set up an integration test with the pandas nightly to
catch upstream issues early.



